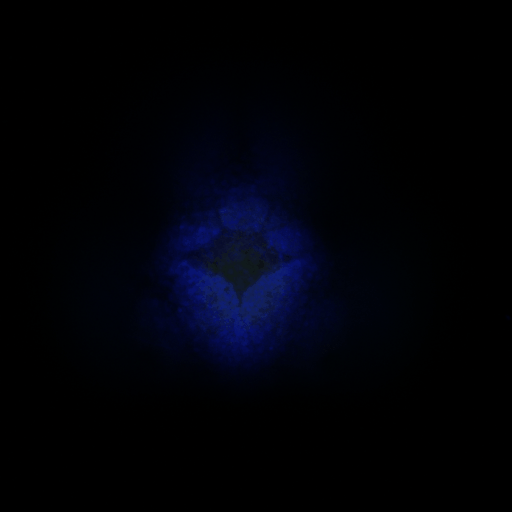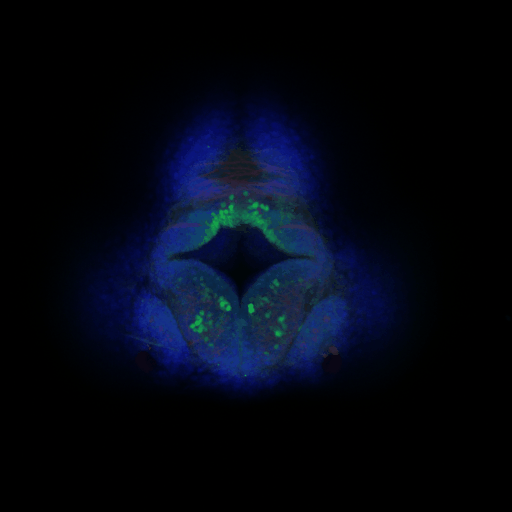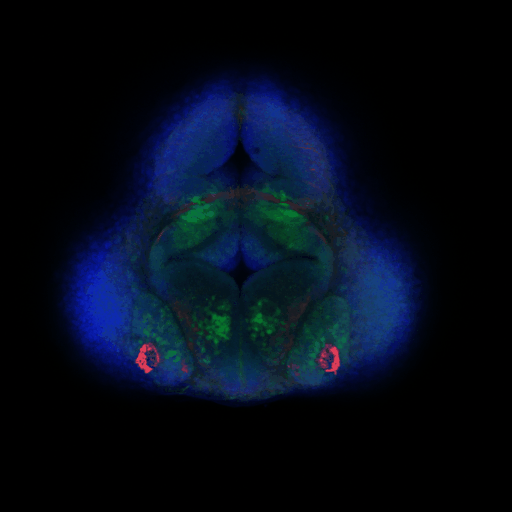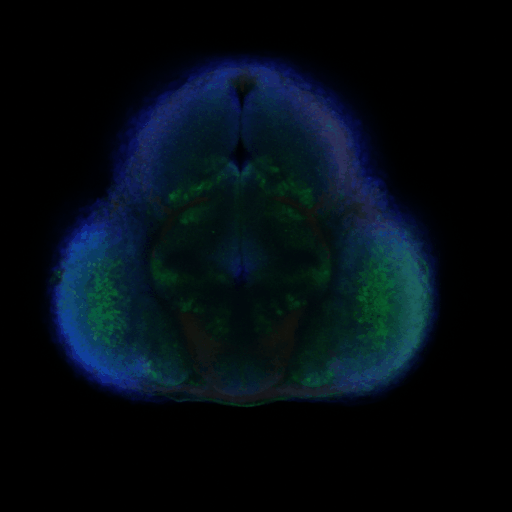
The CeNT’s Biological Imaging Core Facility is a newly established specialised laboratory dedicated to visual examination and analysis of a range of biological processes on a number of different biological systems. Currently, the lab is equipped with an upright laser scanning confocal microscope (Zeiss LSM700).
|  |
|  |
|  |
Research projects at CeNT UW realized at the Biological Imaging Core Facility: | |
Laboratory of Molecular Basis of Synaptic Plasticity led by Magdalena Dziembowska | 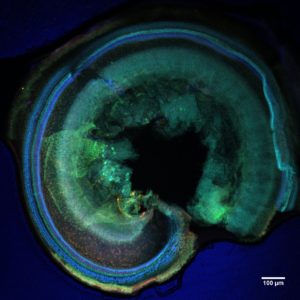 |
Laboratory of Molecular and Cellular Signaling led by Paweł Niewiadomski | 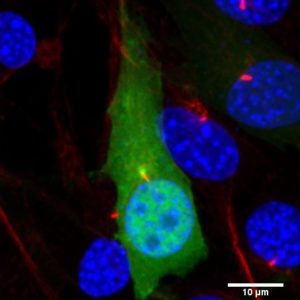 |
Laboratory of Molecular Neurobiology led by Marta Barbara Wiśniewska | 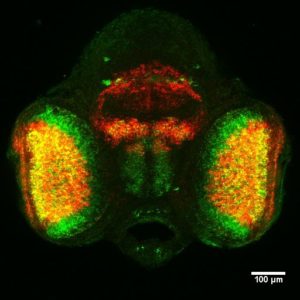 |
1. What is the maximum scan penetration depth of my sample?
It very much depends on the lens used and its effective working distance (WD).
Low power dry objectives (10x or 20x) have longer working distance (5.2 mm and 0.55 mm, respectively) and thus allow for deeper penetration in a range of hundreds of microns but compromising resolution. High power objectives (40x and 63x oil immersion with WD of 0.21 mm and 0.19 mm, respectively) have much thinner scan penetration depth in a range of tens of microns but with high resolution.
2. What is the minimum/maximum thickness of my sample?
It is generally recommended to prepare thin samples (up to 100 μm) for high resolution confocal imaging. If your sample is thicker it will be still possible to scan it on the system but with more laser scattering and thus lower resolution.
3. Is your system suitable for live imaging?
Unfortunately, our confocal system is not equipped with a dedicated environmental chamber which limits the possibility of live imaging of biological specimens.
4. What laser lines are available for fluorescence excitation?
The system comes with 4 diode laser lines: 405nm (5mW), 488nm (10mW), 555nm (10mW), 639nm (5mW)
5. What are the possible experimental applications of your system?
Our system allows for a number of different applications including 3D visualisations and reconstructions, time lapse experiments combined with photoactivation/photobleaching e.g. FRAP or FRET, channel unmixing for clear spearation of overlapping peaks, co-localisation analyses, spectral imaging (lambda scanning) etc. You can find some examples of recent applications under the „Research” tab.
6. What is the maximum resolution you can achieve with your system?
The maximum theoretical spatial resolution in laser scanning confocal microscopy is diffraction-limited according to the following equation:
Resolution (r) = 0.61λ/NA, where λ – imaging wavelength, NA – numerical aperture of the objective
In practical terms, a resolution limit of ~200-300nm in xy and ~700-800nm in z can be achieved using a short laser wavelength of 405nm. This can be further affected by other factors such as sample thickness, sample density, sensitivity to photobleaching etc.
7. What are the microscope scanning modes?
The microscope works in two basic modes – epifluorescence and reflection.
In epifluorescent mode, the laser excites fluorescence within the sample – this is either natural fluorescence (autofluorescence) or a fluorescent dye that has been attached to a specific site on the sample using an antibody label.
In reflection mode the laser light is simply bounced off the surface of the specimen allowing for additional information from specimens that reflect light or have significant fluctuations of refractive index at certain boundaries e.g. to study various cell-matrix interactions.
8. What is the maximum size of a sample?
As long as the sample fits on a single slide it can be imaged even on a macro scale by automatic tiling of individual frames. The stage of our system accommodates up to 10 individual microscopic slides which can be all loaded at the same time.
9. What is the minimum step size of your stage?
Our stage is fully motorised with a travel range in XY of 225 x 85 mm and a minimum step size in Z of 10nm.
10. How do I need to prepare my samples prior to my confocal session?
Your sample preparation protocol is very much dependent on the sample type. It is recommended to mount it on a standard microscopic slide (76x26mm) with a coverslip on top. The samples can be mounted in a solution buffer or other media e.g. glycerol or glycerine and ideally should be immobilised to avoid unwanted movements during acquisition. For more tips and tricks please fill the form in the „Client Zone” section.
11. What is the average time for generating results for commercial samples?
The time depends on the number of samples and experiment type. Typically, a high resolution Z stack acquisition of a 50um thick sample takes up to 1 hour including microscope setup. A half-day confocal session is enough to scan up to 5 samples with high resolution.
12. Can we set a non-disclosure agreement for my commercial research?
Yes, that is possible.
13. How do I get access to my data after finishing my session?
Your data are saved as raw .czi files and will be backed up on the CeNT’s server after each session. They can be then downloaded offline to your memory drive from dedicated CeNT’s workstations.
14. What is the average throughput time needed to process a batch of samples per week?
Depending on the experiment type, we can image and process approximately 10-20 samples per week but this is all arranged on an individual basis once we know more details of the research needs.
15. Is it possible to use your system for a collaborative project?
Absolutely. We are an open access facility and welcome any potential collaborators to conduct their experiments at CeNT UW. Such visitors (if external to UW) will be charged a preferential academic rate compared to fully commercial users. For a full price list please refer to the „Pricelist” tab.
16. What is the average waiting time to analyse my samples?
The microscope is available to all interested users and as such can be quite busy from time to time. However, the average time to book a session should be less than 2 weeks. You can check the current reservation status in the „Client Zone” tab.
17. Is it possible to discuss my research needs or sample preparation protocol before booking an instrument time?
Certainly yes. It is our priority to understand your needs or assist you in a sample preparation step prior to actual microscopy time. We allow our users to fully describe their requirements by filling in an online form in the „Client Zone” tab. This can serve as a good starting point for some face-to face discussions before deciding on the experimental time and post-processing data analysis.
| Internal users (unassisted use) | 32 PLN/h | 8 €/h | 10 $/h |
| External academic users (assisted use)* | 150 PLN/h | 40 €/h | 45 $/h |
| External non-academic users (assisted use)* | 225 PLN/h | 55 €/h | 60 $/h |
Prices include tax.
* For Zeiss LSM 700 Confocal microscope only
A formal acceptation of the rules and regulations is required prior to granting access to core facilities equipment and staff consultations. All information about them you can find in the form below:

| Zeiss LSM 700 |  |
| pon. | wt. | śr. | czw. | pt. | sob. | niedz. |
|---|---|---|---|---|---|---|
1 | 2 | 3 | 4 | 5 |
||
6 | 7 | 8 | 9 | 10 | 11 | 12 |
13 | 14 | 15 | 16 | 17 | 18 | 19 |
20 | 21 | 22 | 23 | 24 | 25 | 26 |
27 | 28 |
|||||
| Olympus IX73 |  |
| pon. | wt. | śr. | czw. | pt. | sob. | niedz. |
|---|---|---|---|---|---|---|
1 | 2 | 3 | 4 | 5 |
||
6 | 7 | 8 | 9 | 10 | 11 | 12 |
13 | 14 | 15 | 16 | 17 | 18 | 19 |
20 | 21 | 22 | 23 | 24 | 25 | 26 |
27 | 28 |
|||||
| Nikon Eclipse fluorescence microscope |  |
| pon. | wt. | śr. | czw. | pt. | sob. | niedz. |
|---|---|---|---|---|---|---|
1 | 2 | 3 | 4 | 5 |
||
6 | 7 | 8 | 9 | 10 | 11 | 12 |
13 | 14 | 15 | 16 | 17 | 18 | 19 |
20 | 21 | 22 | 23 | 24 | 25 | 26 |
27 | 28 |
|||||
Internal User Order Form to be completed and signed by internal CeNT/UW users prior to making reservation assignments.
Formularz zamówienia wewnętrznego użytkownika
External User Order Form to be completed and signed by external academic or external non-academic users prior to making reservation assignments.
If you are a certified user, log in directly via -> Platforma rezerwacji
If you are not a certified user, please complete the form below: