Profile
Team
Services
Regulations
Order forms
Equipment
Contact
Collaboration guide |
Profile
The CeNT’s Genomics Core Facility is a highly specialized laboratory dedicated to analysis of RNA and DNA sequence, structure and interactions in biological and biochemical processes. GCF is registered with a Research Resources ID (RRID): SCR_022718. We employ highly qualified specialists with a broad range of experience and skills. Our lab is equipped with the most up-to-date apparatus including NovaSeq 6000 next generation sequencing system as a core.
Our aim is to enrich academic society with an equal access to a wide spectrum of highly specialized instruments for research in Genomics and its all related disciplines, such as Metagenomics, Transcriptomics, Epigenomics, Interactomics, Oncogenomics, Structural Genomics and many other dynamically developing fields. The goal is achieved by professional support in planning of experiments, its conduction and results analysis, so that even inexperienced users may benefit from modern techniques in molecular biology and bioinformatics. At the same time, by collaboration with non-academic entities we hope to build a bridge between research and application and to initiate a wider use of the achievements of modern genomics in industrial or medical practice.
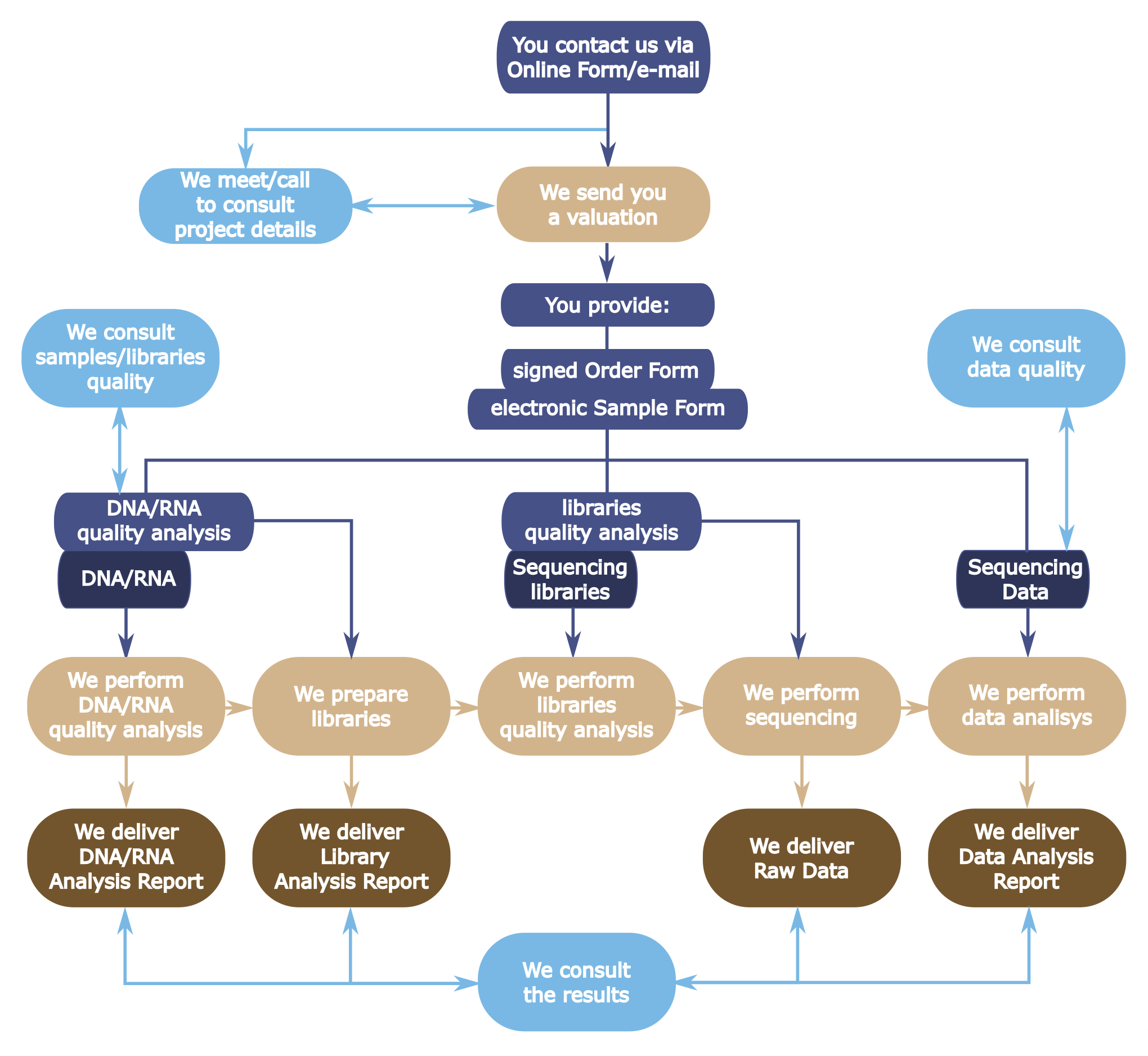
To find out what we can do for you please read about services we provide, how we can collaborate and contact us via on-line form. Also do not forget to read the Regulations on-line form/ |
|
Services
Next Generation Sequencing on the Illumina machines is a three-part process that consists of sample preparation (sequencing libraries construction), sequencing run and data analysis. The three services may be priced and provided either as a whole or separately. Different rates apply to the following user groups: internal academics (CeNT and UW), external academics and commercial collaboration. Please contact us for pricing. Additional services on request are also available. |
|
Equipment
| Illumina NovaSeq 6000
The very high-throughput, the most cost-effective and flexible sequencer in Illumina portfolio.
- High scalability:
- 65-3000 Gb/run
- 800M-10B reads/run
- up to 2×250 bp read length
- Very high quality (typically ≥ 95% of bases higher than Q37 at 2×100 bp)
- High speed of sequencing: 13-48 h
- Can run two flow cells in parallel at once
|
 |
| Illumina NextSeq 500
The mid-throughput sequencer for a wide range of application.
- Scalability:
- 20-120 Gb/run 130-400M
- reads/run
- up to 2×150 bp read length
- High quality (typically ≥ 90% of bases higher than Q30 at 2×75bp)
- High speed of sequencing: 11-29 h
|
 |
| Illumina MiSeq
The relatively low-throughput sequencer for small projects, amplicon or test sequencing runs.
- High scalability of low-throughput runs:
- 0.3-15 Gb/run
- 1-25M reads/run
- The longest read length: up to 2×300 bp read length
- High speed of sequencing: 6-56 h
|
 |
| Systems for Automatic Reaction Preparation |
|
| Freedom EVO 150/8
Automatic pipetting station for reaction set up.
- Two types of robotic arm: 8 – tip pipetting arm (LiHA) and labware movement arm (ROMA)
- Scalability with multiple format and size of robotic carriers – from 384-well plates to reservoirs for hundreds of ml
- Wide range of modules for liquid manipulation and carrying out reactions
- Heating system
- Fluorometric system
- Magnetic separation module
|
 |
| Nucleic acids quality and quantity assessment |
|
| TapeStation 2200
Nucleic acid and proteins quality control automated system based on electrophoresis on tapes
- High sensitivity – from 10 pg for DNA , 200 pg for RNA, 5 ng for protein
- High resolution – up to 5 bp for DNA/RNA and 0.5 kDa for protein
- Broad range of different reagents for:
- DNA (from 30bp to genomic)
- total RNA and mRNA
- protein (up to 200 kDa)
- High flexibility – from one sample at a time
|
 |
| BioAnalyzer 2100
Nucleic acid quality control automated system based on electrophoresis on chips
- Very high sensitivity – from 5 pg for DNA , 50 pg for RNA, 0.3 ng for protein
- High resolution – up to 5 bp for DNA/RNA and 0.5 kDa for protein
- Broad range of different reagents for:
- DNA (from 30 bp to 10 kbp)
- total RNA and mRNA
- small RNA
- protein (up to 250 kDa)
- 11-12 samples at a time
|
 |
| LightCycler 480
Real-time PCR system
- High-throughput system – 384-well plate format
- Nucleic acids quantity and quality assessment for:
- relative expression analysis
- genotyping
- sequence specific detection
- PCR amplification optimization
- sequencing libraries relative concentration measurements
|
 |
| Nucleic acids preparation |
|
| Covaris S220
Ultra-focused, very precise sonication system with cooling
- Very high repeatability assured by:
- ultra-focused sound wave
- external cooling system
- water degassing
- vessel construction
- High precision of very equal physical fragmentation to particular size of DNA
- High efficiency of cell disruption and chromatin fragmentation
|
 |
| Covaris M220
Ultra-focused, very precise sonication system
- Very high repeatability assured by:
- ultra-focused sound wave
- internal temperature control system
- vessel construction
- High precision of very equal physical fragmentation to particular size of DNA
- High efficiency of cell disruption and chromatin fragmentation
|
 |
|
|
Contact
To facilitate the transfer of all the information needed to start cooperation with us, we have prepared a form for you. When initiating contact, we encourage you to complete it.
Contact form
Questions and information not covered by the form should be provide by e-mail or by phone.
Samples preparation and sequencing
Dorota Adamska, d.adamska@cent.uw.edu.pl, +48 22 55 43628
Bioinformatic analysis
dr Krzysztof Goryca, k.goryca@cent.uw.edu.pl, +48 22 55 43681 |