|
Laboratory The CeNT’s Genomics Core Facility is a highly specialised laboratory dedicated to analysis of RNA and DNA sequence, structure and interactions in biological and biochemical processes. GCF is registered with a Research Resources ID (RRID): SCR_022718. We employ highly qualified specialists with a broad range of experience and skills. Our lab is equipped with the most up-to-date apparatus including NovaSeq 6000 next generation sequencing system as a core. All to achieve high quality service, constant development and to conduct significant contribution to all our cooperations!
|
|
GCF Mission Our aim is to enrich academic society with an equal access to a wide spectrum of highly specialized instruments for research in Genomics and its all related disciplines, such as Metagenomics, Transcriptomics, Epigenomics, Interactomics, Oncogenomics, Structural Genomics and many other dynamically developing fields. The goal is achieved by professional support in planning of experiments, its conduction and results analysis, so that even inexperienced users may benefit from modern techniques in molecular biology and bioinformatics. At the same time, by collaboration with non-academic entities we hope to build a bridge between research and application and to initiate a wider use of the achievements of modern genomics in industrial or medical practice. To find out what we can do for you please read about services we provide, how we can collaborate and contact us via on-line form!!! Also do not forget to read the Regulations on-line form!!! |
| PhD Jeffrey Palatini - head of Genomics Core Facility | |
| PhD Eng. Dorota Adamska - experiment design consultant, sample preparation and sequencing platforms operator Research qualifications and technical skills
|  |
| PhD Krzysztof Goryca - bioinformatic analysis Research qualifications and technical skills
| |
| MSc Michał Szeląg - sample preparation and instrument operator Research qualifications and technical skills
|  |
| PhD Julia Rachowka - sample preparation and instrument operator Research qualifications and technical skills
|
Genomics Core Facility works on the most up-to-date apparatus for nucleic acids manipulation and analysis. We strive to make able to take advantage of the equipment to all interested research group, serving full support in operation and substantive help in the preparation of material for analysis. If interested please contact as via on-line form.
Next Generation Sequencing Systems
Illumina NovaSeq 6000
|
The very high-throughput, the most cost-effective and flexible sequencer in Illumina porfolio
|
Illumina NextSeq 500
|
The mid-throughput sequencer for a wide range of application
|
Illumina MiSeq
|
The relatively low-throughput sequencer for small projects, amplicon or test sequencing runs
|
Systems for Automatic Reaction Preparation
Freedom EVO 150/8
|
Automatic pipetting station for reaction set up
|
Nucleic acids quality and quantity assessment
TapeStation 2200
|
Nucleic acid and proteins quality control automated system based on electrophoresis on tapes
|
BioAnalyzer 2100
|
Nucleic acid quality control automated system based on electrophoresis on chips
|
LightCycler 480
|
Real-time PCR system
|
Nucleic acids preparation
Covaris S220
|
Ultra-focused, very precised sonication system with cooling
|
Covaris M220
|
Ultra-focused, very precised sonication system
|
Price list for NGS services
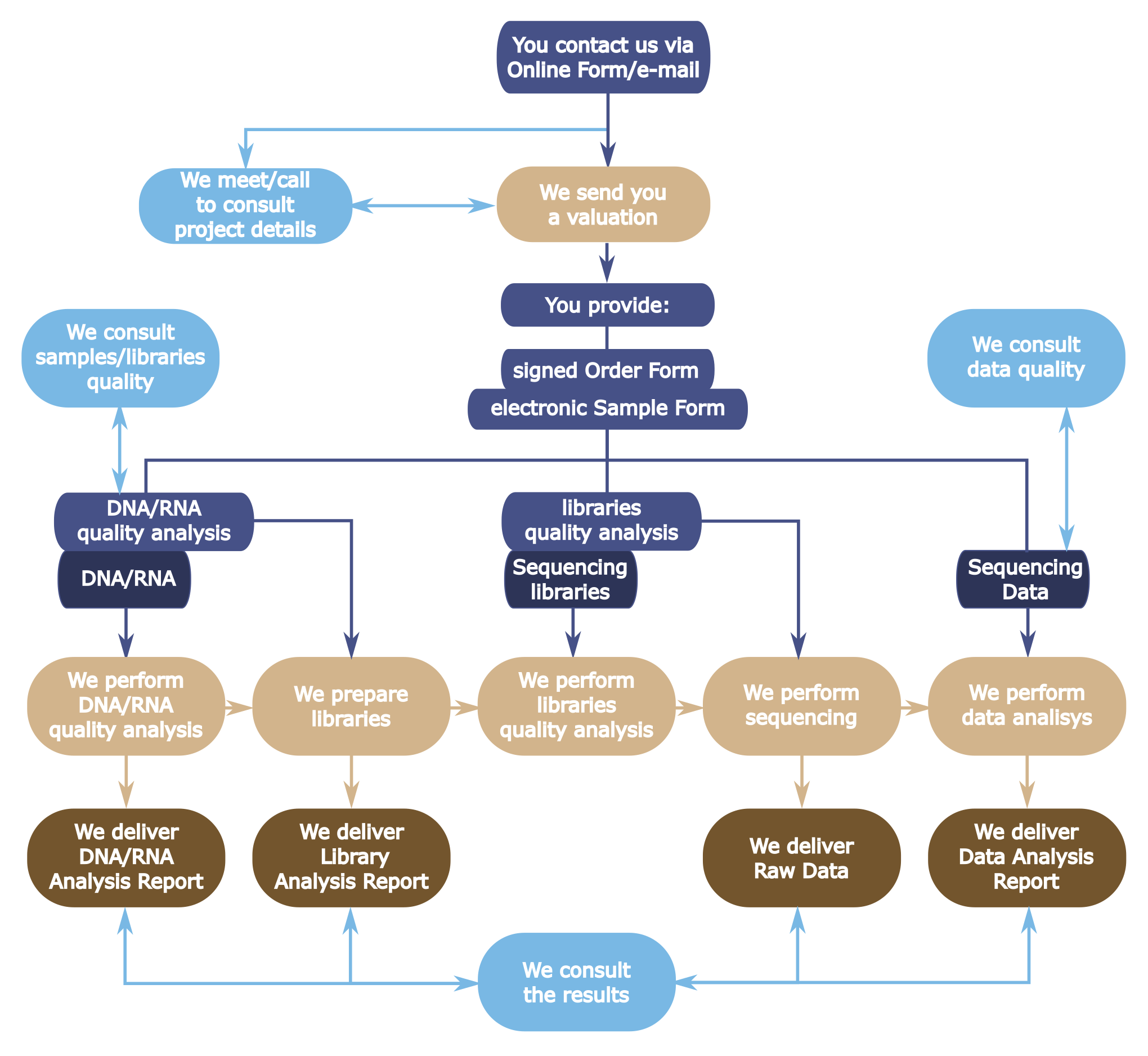
Next Generation Sequencing on the Illumina machines is a three-part process that consists of sample preparation (sequencing libraries construction), sequencing run and data analysis. The three services may be priced and provided either as a whole or separately. Different rates apply to the following user groups: internal academics (CeNT and UW), external academics and commercial collaboration. Please contact us for pricing. Additional services on request are also available.
Cooperation or orders in Genomics Core Facility CeNT UW is equivalent to acceptance of the following rules. Please read the regulations before starting the consultation and starting work.
General rules for the use of the CeNT UW
Genomics Core Facility
I. General provisions
- The equipment is localized in room 03.231 and 05.101-05.115 in the CeNT I building.
- Genomics Core Facility personnel:
Jeffrey Palatini (head) (e-mail: j.palatini@cent.uw.edu.pl);
Sample Processing Section:
Dorota Adamska (e-mail: d.adamska@cent.uw.edu.pl);
Michał Szeląg (e-mail: m.szelag@cent.uw.edu.pl);
Bioinformatics Section:
dr Krzysztof Goryca (e-mail: k.goryca@cent.uw.edu.pl); - Direct use of the equipment is restricted to the personnel of the Genomics Core Facility.
- Users are requested to submit a brief description of the planned experiment and/or data analysis proposal by filling an electronic form. The project description is the basis for consultations scheduling, obtaining acceptance for the experiment from the Sample Processing Section and/or Bioinformatics Section, and enables them to plan sequencing and data analysis, respectively.
- Users are requested to accept the preliminary cost estimate before starting the sample processing or data analysis.
- Users are obliged to familiarize themselves with these rules.
II. Project submission
- The submissions of experiment and data analysis proposals are made through the Genomics Core Facility website:
“https://cent.uw.edu.pl/en/core-facilities/genomics-core-facility/” - By accepting the terms and conditions the Users declare that the samples and data provided have not been obtained in violation of applicable law, as well as their further processing requested by the Genomics Core Facility personnel is fully legal.
III. Sample preparation
- Users submitting DNA/RNA samples must provide nucleic acid quality and quantity data or request such an analysis to be carried out by the Genomics Core Facility. Samples that do not meet quality requirements as specified by the selected sequencing library preparation protocol will be processed only upon written consent, wherein the User agrees that no guarantee is given for the successful library construction and the full costs of the library construction attempts (including the unsuccessful ones) are to be fully covered by the User.
- Users submitting DNA libraries are required to follow sample preparation protocol approved by the head of the Sample Processing Section. They must also present data on the quality of the libraries delivered (results from Agilent Bioanalyzer or equivalent instrument) or request the quality analysis to be carried out by the Genomics Core Facility. Any modification (e.g. resulted from a human error) should be reported before the libraries are handed over for sequencing. The Genomics Core Facility bears no responsibility for the quality of results obtained, based on sequencing User-delivered libraries.
- Users submitting sequencing data for analysis should deliver data in BCL, FASTQ, or BAM format.
IV. Provided services
- The services are rendered, based on the initial consultation (see Collaboration Guide) and according to the written cost estimate provided to and accepted by the User.
- The Genomics Core Facility personnel is responsible for scheduling the services. Generally, User requests are being processed on a first come, first served basis. However, the personnel reserves the right to queue the samples provided, according to the availability of measurement time and sequencing consumables, unless the User agrees to bear the full costs of the sequencing process, as estimated by the Genomics Core Facility personnel.
- The general services offered by the Genomics Core Facility include:
- Consulting services (help with designing the experiment, customizing sample preparation protocols, data interpretation);
- Sample processing (eg. DNA/RNA quality assessment, DNA library preparation);
- Sequencing run set up and raw data preprocessing (FASTQ generation, demultiplexing and basic quality report);
- Data analysis (basic processing as well as tailored analysis and data visualization are available).
- The services rendered are described on the Genomics Core Facility website and individually adjusted in accordance with experiment-specific modifications of the standard protocols and additional services requested (e.g. sample quality control, data analysis).
- A consultation is expected to precede any sample processing and/or data analysis (see Collaboration Guide). As a result of the consultation, an optimal analytical strategy is agreed upon and the estimated cost of services (calculated based on the Pricelist) is provided in a written form and explained.
- The Genomics Core Facility personnel reserves the right to reject requests for the provision of services in objectively justified cases. In the event of a refusal, a constructive feedback will be provided to both the User who had requested the service, and to the appropriate head of the respective laboratory (where applicable).
V. Libraries storage, data storage & security
- The results will be made available to the User in an adequate form::
- Report on the methodology and work results (for Sample processing service);
- Raw, demultiplexed data with basic quality report (for Sequencing service);
- Full report with a summary addressing the User’s expectances (for data analysis service).
- Users are expected to collect data within 14 days from the time the results became available to them. After that time, access to the data will require intervention of the head of the lab, from which the project originated.
- The samples and libraries remained will be stored for 6 months and provided to the User on demand or may be subjected to further sequencing on separate sequencing request.
- Raw data will be stored on a server for a duration of 6 moths. On request, they will be made available to the User.
- Additional storage time of libraries or raw data requires written request from the User within 6 months from making results available to them and may result in storage cost covering by the User.
VI. Publication of data
- Acknowledgment: For basic analyses, where a minimal intellectual contribution from the Genomics Core Facility personnel has been requested, their work should be clearly identified in the acknowledgments section of resulting publications with marked Research Resources ID (RRID:SCR_022718). Sequences and data analysis reports will be provided along with a generic statement about the instrumentation and the software utilized.
- Co-authorship: When a significant intellectual contribution from the Genomics Core Facility personnel has been requested and delivered, their work must be acknowledged by naming them as co-authors of resulting publications. This is a default mode of agreement for projects involving the development of new sequencing and data analysis approaches and methods or projects requiring complex data processing and/or mining. A selection of publication-ready figures and an input into writing of the relevant sections of the paper will be provided by the Genomics Core Facility personnel.
Collaboration guide
The method of contacting us can be found in the Contact tab, and all the necessary documents in the Attachments tab.
Internal User Order Form to be completed and signed by internal CeNT/UW users prior to making reservation assignments.
External User Order Form to be completed and signed by external academic or external non-academic users prior to making reservation assignments.
To facilitate the transfer of all the information needed to start cooperation with us, we have prepared a form for you. When initiating contact, we encourage you to complete it.
Contact Form
Questions and information not covered by the form should be provide by e-mail or by phone:
Samples preparation and sequencing
Dorota Adamska (e-mail: d.adamska@cent.uw.edu.pl), +48 22 55 43628
Bioinformatic analysis
dr Krzysztof Goryca (e-mail: k.goryca@cent.uw.edu.pl), +48 22 55 43681