| Project Leader: Agata Perlińska, MSc | Project period: 2019 - 2021 |
| Project funding: PRELUDIUM 15, NCN | |
| Project description: My project focuses on the study of complex proteins that possess a knot-like fold in their structure (Figure 1). This type of proteins was first observed over 20 years ago, and now protein databases hold over a thousand solved knotted structures with different types of entanglements. Knotted proteins make a new and exciting area of study that brings together many disciplines ranging from biology, through biochemistry, biophysics up to mathematics. In everyday life, we often use knots for stabilization and it would be natural to expect that molecular knots will be present in proteins with structural function. However, the majority of them are found in enzymes. Thus, the questions occur: whether a knotted region helps the protein in catalyzing the chemical reaction? Does it have a specific role to maintain? Especially, since we already know that this function can be performed without a knot, although through a different mechanism.
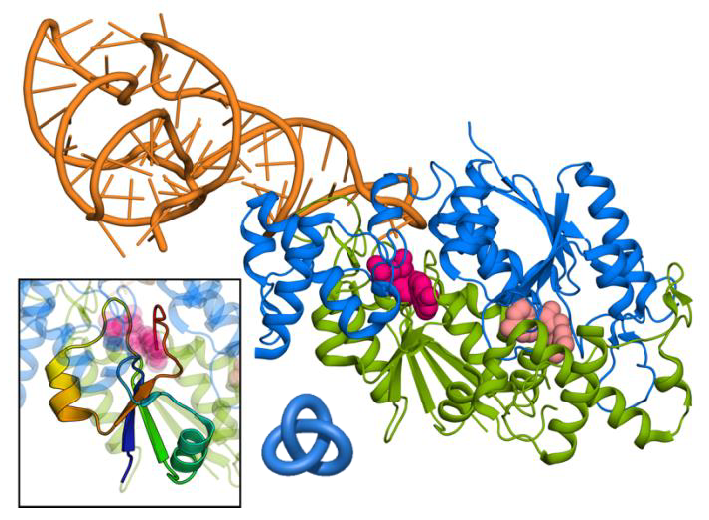
Figure 1: Cartoon representation of TrmD protein (subunits colored with blue and green) in a complex with tRNA (orange) and 2 ligands (pink). Inset shows the knotted region of the protein. To shed light on these, fundamental for structural biology, questions, this project focuses on both understanding what is the mechanism of function of knotted proteins and what is the role of knot in it. The project is based on proteins responsible for methylation of nucleic acids. In particular, on bacterial methyltransferase TrmD – a protein with the simplest and most commonly found type of knot (Figure 1 inset). TrmD’s activity depends on the magnesium concentration in the cell – Mg2+ ion is essential for the efficient catalysis. The protein is a vital enzyme made up of two identical subunits and so with two identical active sites. However, the protein exhibits ‘halfof-the-sites’ activity – only one site is active at a time. Since knotted region creates the crevice for the ligand to bind, it implicates the role of the knot in the asymmetrical behavior of the protein. Analysis of the available crystal structures of different complexes of TrmD does not yield an explanation to its mechanism of the function – neither the complex is asymmetric nor the magnesium ion is present. However, crystallization gives only a static view of the structure, and the answer must lie in the dynamic behavior of the protein, which is hard to analyze experimentally. Therefore, the main aim of this project is to determine and characterize, using theoretical tools, proper conditions and behavior of the knotted protein that leads to its activation. My idea of solving the question of the role of the knot is based on combined approach of molecular dynamics simulations and quantum chemical calculations with the support of the experimental results. I will perform several simulations that show the behavior of the protein in water (conditions similar to those naturally occurring in the cell) in atomistic detail. Comparison of simulations with different number of ligands, different starting structures, and different conditions (presence/absence of magnesium ions) will allow me to capture the changes that induce the protein into the activation state. In particular, how the asymmetry is propagated in the protein, and what is its role. Also, how the magnesium influences the structure, and whether its importance in catalysis is correlated with its influence on asymmetry. All this information would also show if the knotted region is a part of the activation mechanism of TrmD protein, and hopefully point to its role in it. The final step of the project is to check whether found features apply to other knotted proteins (in particular other methyltransferases). Specifically, with aid of the quantum calculations I will determine if the magnesium is an important part of catalysis of methyl transfer in those proteins, as it is for TrmD. The study of knotted proteins, and TrmD in particular, is not only a basic research but also a step towards real-life applications, such as creation of novel antibiotics. World Health Organization recently published a list of 12 drug-resistant bacteria that should be treated as high-priority pathogens. TrmD, a vital and strictly bacterial protein, present in all of these strains, is thus an excellent antimicrobial target for drug design. Therefore, the results of this project will also bring information crucial for finding a new, potent drug. Success of this interdisciplinary project requires vast knowledge from many fields. My background in biology, bioinformatics and biophysics, and my supervisor’s (Joanna Sulkowska) many years of experience with knotted proteins, as well as collaboration with an experimental group headed by prof. Hou, makes me uniquely qualified to complete it. |
|
|
Interdisciplinary Laboratory of Biological Systems Modelling |
|