
Research Focus
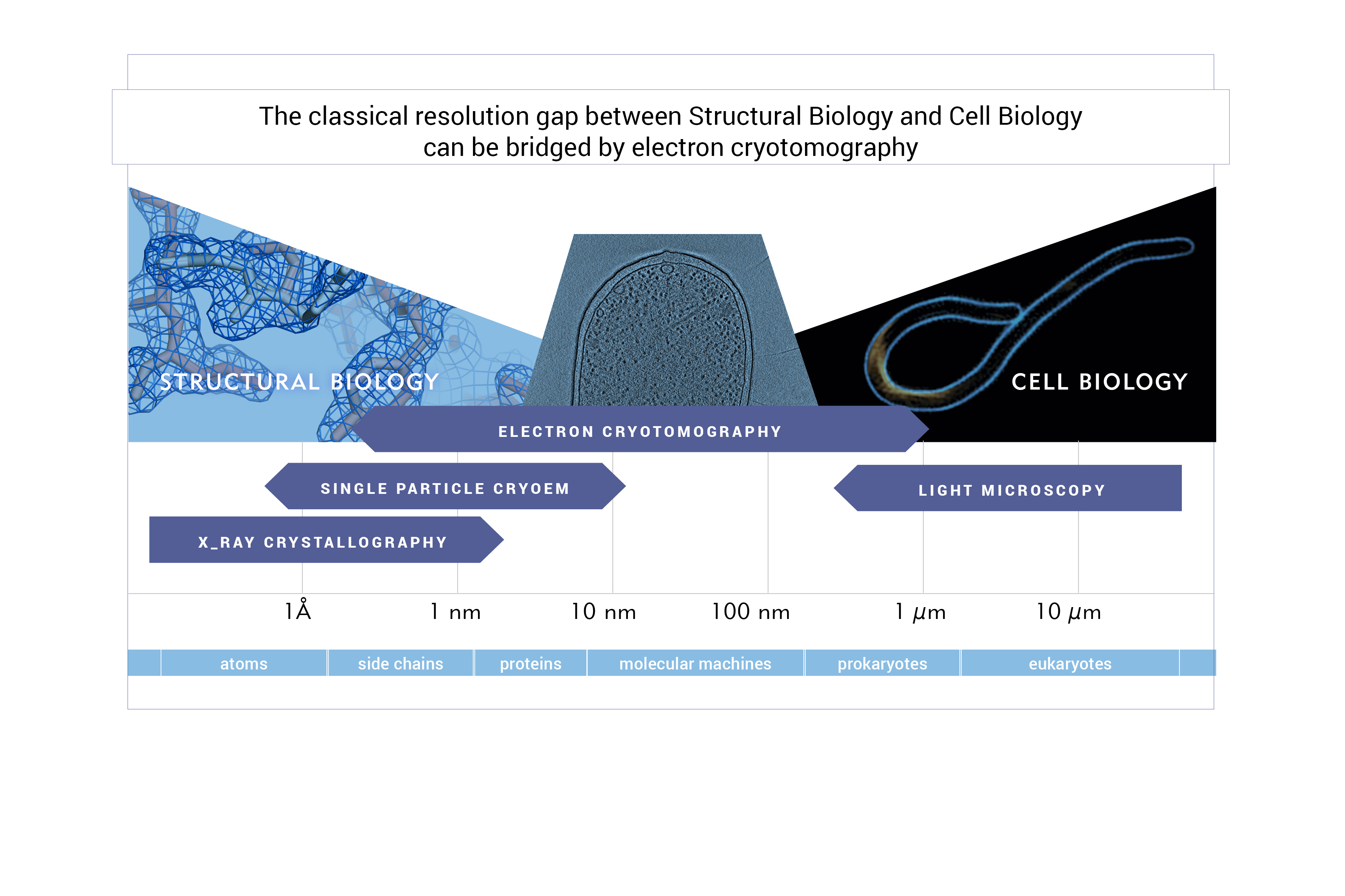
Our research aims to understand the organisation and architecture of cells at the molecular level. We utilize a multiscale approach, combining imaging across scales (X-ray protein crystallography and fluorescence light microscopy bridged by electron cryotomography) with biochemistry (in vitro reconstitutions).
Our biological focus is on explaining certain aspects of archaeal structural cell biology. The Archaea have been described as a major domain of life that share biological traits with both the Bacteria and the Eukarya: the archaeal information-processing system is common with the Eukarya, whereas the metabolism is common with the Bacteria. We find the Archaea fascinating for two reasons. Firstly, members of this domain of life frequently thrive in extreme environments, and it is of great interest to find out how their cellular ultrastructure, including the liquid-liquid phase separated membraneless compartments, facilitate survival in adverse environmental conditions. Secondly, it has recently emerged that the Archaea encode for eukaryotic signature proteins involved in membrane bending, vesicle biogenesis, and trafficking, and display an elaborate internal membrane organization. We believe that understanding the cell biology of the Archaea in the context of eukaryogenesis is a prerequisite to decipher the morphological transition that characterised the evolution of the complex eukaryotic cellular organisation.
The understanding of this process is essential not only for evolutionary studies probing the origins of life but also for fundamental biomedical research. In particular, liquid-liquid phase separation is a widespread process in human cells. Many phases separating proteins contain prion-like domains and aberrant phase-transitions result in the formation of insoluble amyloid fibrils, like those associated with neurodegenerative diseases in mammals. I strongly anticipate that investigating liquid-liquid phase separation in Archaea, close prokaryotic relatives of Eukarya, will lead to a better understanding of basic principles, regulation and origins of the process. As a result, we will generate a wealth of information relevant to the overall ReMedy goals.
To closely observe archaeal cells in 3D we employ electron cryotomography – an electron cryomicroscopy (cryoEM) technique that resolves unique structures in situ, in a native state, in three dimensions, and at the molecular range, effectively enabling structural studies in intact, frozen-hydrated cells. For cryoEM data collection we will have access to an in-house 200 kV Thermo Fisher Glacios electron cryomicroscope equipped with a Falcon 3EC direct electron detector and a phase plate, and will regularly collect our data on a Thermo Fisher Titan Krios G3i equipped with a phase plate, BioQuantum energy filter and both K3 and Falcon3EC direct electron detectors that is located at the National Centre for Electron Cryomicroscopy in Kraków.
Alvarez-Malmagro, J., Jabłonowska, E., Nazaruk, E., Szwedziak, P., & Bilewicz, R. (2020).
Bioelectrochemistry, 134(107516).
Alvarez-Malmagro, J., Matyszewska, D., Nazaruk, E., Szwedziak, P., & Bilewicz, R. (2019).
Langmuir, 35(50), 16650-16660.
| Title | Deadline for applications |
|---|---|
| Postdoc in ReMedy (Laboratory of Structural Cell Biology) | 15/12/2019 |
| ReMedy – Postdoc in Laboratory of Structural Cell Biology | 10/08/2019 |
| Public Procurement and Financial Specialist | 30/07/2019 |
| ReMedy – Postdoc in Laboratory of Structural Cell Biology | 30/04/2019 |
| PhD Student at ReMedy | 30/04/2019 |



